Environmental DNA, or eDNA, is an exciting and emerging tool that Park scientists are using to gain a greater understanding of local fish biodiversity and population dynamics. Paired with traditional fish survey methods, eDNA can provide a more comprehensive picture of what fish species are present in the River.

In 2019, HRPK’s River Project launched an eDNA survey with partners at Cold Spring Harbor Laboratory. During this two-year study, HRPK staff scientists collected samples in the Park, filtered them and extracted the DNA from the filters. These samples were then sent to experts at Cold Spring Harbor Lab for DNA isolation, next generation sequencing and metabarcoding. Check out the results here!
Today, the Park continues eDNA research with partners from the New York State Department of Environmental Conservation, Lamont-Doherty Earth Observatory and the Center for the Urban River at Beczak to map the presence of fish species of interest within the Lower Hudson River Estuary.
What is eDNA?
DNA is genetic material possessed by all life, and it carries information about how a living thing will look and function. Aquatic eDNA is genetic material from the sloughed cells and feces of living marine organisms. Within the Park’s Sanctuary waters, we can find DNA from a wide range of wildlife, including oysters, seahorses and mallard ducks, all in a small sample of River water. By analyzing this genetic material, we can understand more about biodiversity, migration patterns of fish and other wildlife and monitor species of interest, such as endangered or invasive species, that are present within our Estuarine Sanctuary.
Learn how we collect and process eDNA in HRPK
About the Cold Spring Harbor Lab eDNA Fish Study
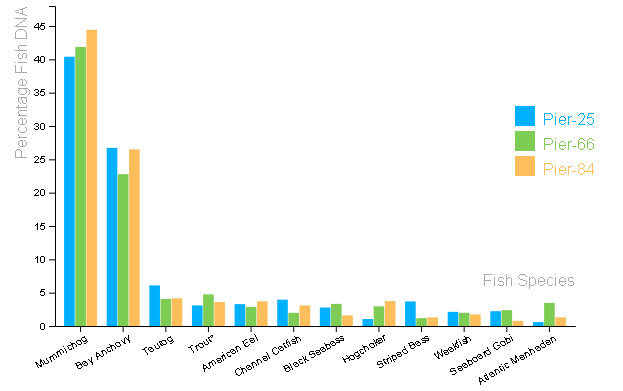
In this study, Hudson River Park staff, interns and local students collected water samples from Hudson River Park’s Pier 25, Pier 66 and Pier 84 (see the chart below). These samples were taken biweekly throughout the year and filtered to collect DNA. Cold Spring Harbor Laboratory then extracted and amplified the desired barcode region, a section of the 12S ribosomal RNA, using polymerase chain reaction (PCR). After confirming the region was copied, the DNA was sequenced using “Next Generation Sequencing.”
The sequencing produces so much data that a sophisticated program is needed to check its quality and analyze the results. The bioinformatics program compares all of the sequences to a fish database, identifying the species of fish and counting the fish DNA sequences for each species.
Interestingly, the two most prevalent DNA matches were those of mummichogs and bay anchovies – species known to the area but that are not seen using our conventional survey methods. This is likely due to the inherent bias in trap sampling which precludes species that behaviorally avoid structures. There were also some species identified that were a bit of a surprise, such as salmon which might have entered our waterways through sewer discharge events. Some of our unexpected species might also live upstream in the Hudson and their DNA flowed downriver, such as largemouth bass and pumpkinseed sunfish. Some common species didn’t show up in our DNA samples, which might have to do with the fishes’ own behaviors (where it swims in the water column) or how the DNA is copied using PCR. It’s possible that the DNA primers might not have bonded well enough due to variance of the target region or that our bioinformatics database doesn’t include those fish species yet.
Overall, this project helped the Park and its partners learn more about the applicability of eDNA in an urban estuarine environment and about local fish dynamics.
Visit this page or follow the link on the chart below for an interactive map highlighting notable eDNA findings from this study. Stay tuned as we share additional data and reports on our findings.